A Cutting-edge Tool used for Investigating the Electrophysiological Properties of iPSC-Derived Neurons and Astrocytes
Multielectrode Array (MEA) Assay
Multielectrode Array (MEA) Assay:
What is a Multielectrode Array (MEA) Assay with human iPSC-derived neurons?
The Multielectrode Array (MEA) Assay is a cutting-edge tool that can be used for investigating the electrophysiological properties of neurons and astrocytes derived from human induced pluripotent stem cells (iPSCs). The MEA system consists of a grid of tightly packed microelectrodes, often referred to as an electrode array or multielectrode array, embedded at the bottom of specially designed MEA plates. These microelectrodes record the electrical activity of neurons, facilitating real-time, non-invasive monitoring of neuronal network function over extended periods. Neurons are cultured directly on the MEA electrodes, and the spontaneous or stimulus-evoked electrical signals they produce are detected and amplified by the underlying electrodes, enabling MEA recording of network activity.
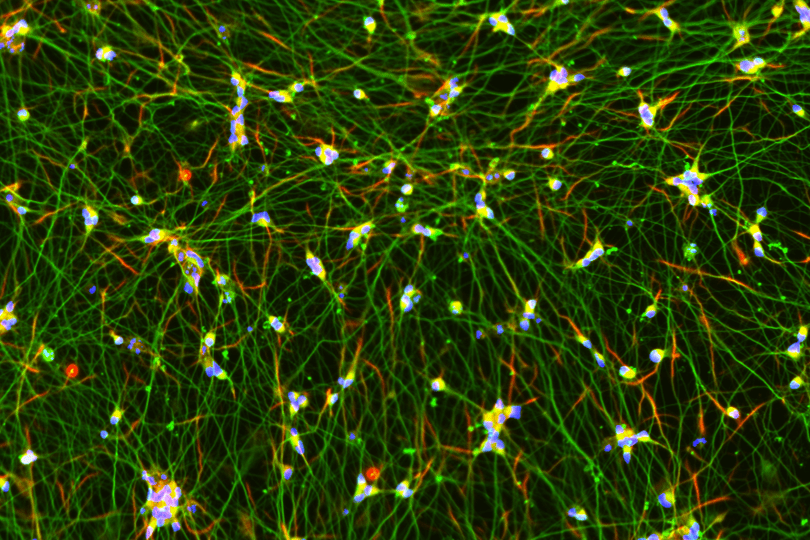
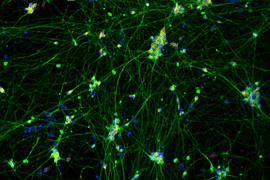
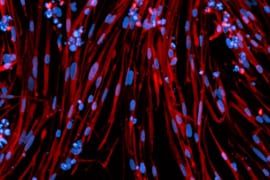
MEA recording of Quick-Neurons™ Excitatory and primary astrocytes on Axion Maestro MEA plate after following this application protocol
MEA options
MEA platforms can vary in their configurations, including electrode density, providing different capabilities for neuronal network analyses.
Traditional platforms:
Traditional low-density electrode arrays offered by companies like Axion Biosystems, Alpha-MED Scientific, and Multi Channel Systems provide an entry point into MEA research. These systems offer fewer electrodes per well (usually 16 to 64), making them well-suited for examining overall network activity and basic neuronal functionality. These plates, made of transparent plastic or glass, allow for observing culture changes over time with a typical inverted microscope. Despite providing a higher throughput format, these traditional plates often lack enough electrodes to cover the entire well, requiring the careful placement of cells or well-optimized culture conditions.
Newly emerging platforms:
High-density MEA platforms, such as those developed by MaxWell Biosystems and Sony, contain hundreds to thousands of electrodes per well by utilizing complementary metal-oxide semiconductor (CMOS) technologies. These systems offer much higher spatial resolution, allowing researchers to examine the activity of individual neurons and intricate neuronal network interactions at a subcellular level. However, these newer technologies have some limitations. They generally lack higher throughput plate formats and the plate bottoms are not transparent, preventing observation of cell morphology during culture with a typical inverted microscope.
Mono-culture vs. Co-culture:
The choice between mono-culture and co-culture depends on the specific research goals and requirements. A mono-culture of iPSC-derived neurons is simpler to maintain and allows for a focused study on neurons. However, it doesn't capture the complexity of the brain's multi-cellular interactions and takes time to mature, affecting the frequency of network bursts in MEA recording. In contrast, a co-culture of iPSC-derived neurons with astrocytes and/or microglia better represents physiological conditions, potentially leading to more accurate results. The presence of astrocytes can also accelerate network maturation compared to mono-culture conditions. However, this culture setup is more complex to maintain and interpret due to interactions between different cell types.
Advantages against other alternative assays
MEA assays offer several key advantages over other techniques.
High content imaging:
High-content imaging allows for the visualization of cellular morphology and processes but typically does not provide functional information. MEA assays, in contrast, allow for real-time monitoring of the functional electrical activity of neuronal networks.
Calcium Flux assays:
Calcium Flux assays are another method used to monitor functional neuronal activity from whole wells in a high-throughput manner, but they only provide indirect measurements and often require the use of dyes or genetic modifications. MEA assays provide a non-invasive, direct, and more physiologically relevant measurement of neuronal electrical activity without the need for such modifications. MEA platforms can generally measure more parameters from recordings. Some labs use calcium flux assays for initial screening to narrow down drug candidates, then gain deeper insights with more parameters from MEA assays.
Applications in Drug Discovery
Disease Modeling / Early Discovery:
MEA assays play an important role in various stages of drug discovery. In disease modeling and early discovery stages, MEA assays with iPSC-derived neurons enable the in vitro modeling of neurological diseases. Researchers can study disease phenotypes and screen for potential drug candidates using neurons derived from iPSC generated from patients with specific diseases.
Safety Pharmacology:
In the later stages of drug development, MEA assays are used in safety pharmacology to test the safety of new drug candidates before moving to clinical trials. These assays can help identify potential seizure liability and contribute to the assessment of a compound's safety profile.
How Elixirgen Scientific iPSC Differentiation Technology Could Help
Elixirgen Scientific's iPSC differentiation technology provides high-quality, patient-specific neurons for use in MEA assays. These iPSC-derived neurons retain the genetic information of the donor, allowing for more personalized and disease-specific research. When coupled with MEA assays, this technology significantly enhances our understanding of disease mechanisms, supports the identification of potential therapeutic targets, and accelerates the drug discovery process. The integration of MEA systems and iPSC technology represents a powerful tool in neuroscience research and drug development. Neurons from Elixirgen Scientific have been used in several publications and presentations. Elixirgen Scientific also offers multiple MEA services for drug discovery research groups, providing well-optimized culture conditions to obtain robust datasets.