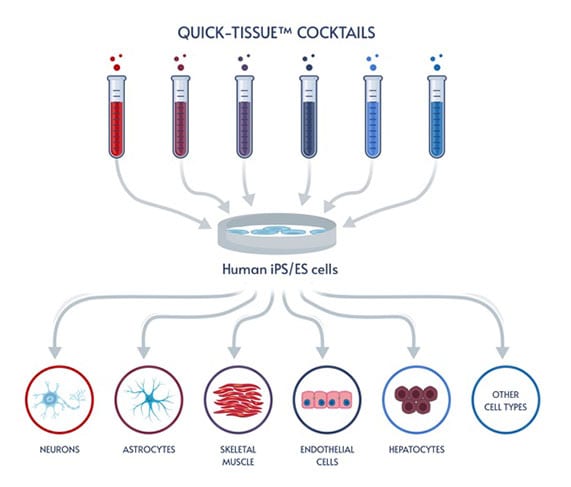
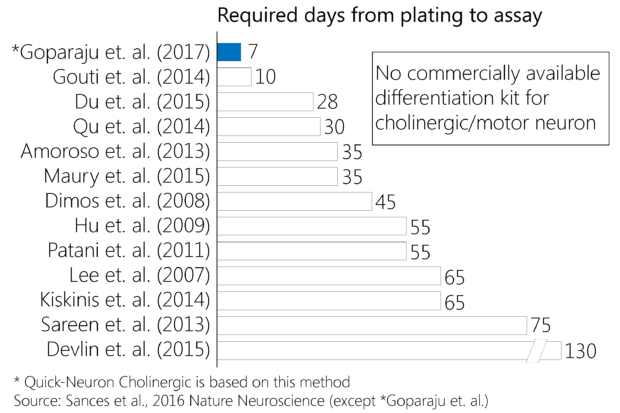
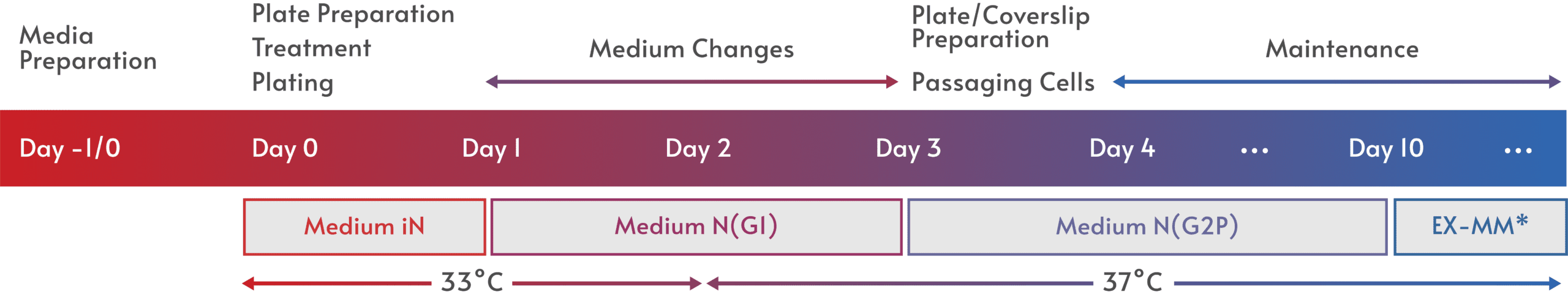
Fast and easy iPSC differentiation enables more effective ways to model human biology and disease, opening the door to a wealth of applications.
Our iPSC Differentiation Technology
The Promise of iPSCs
Human induced pluripotent stem cells (iPSCs) have been expected to drive the biological revolution of research innovation by giving researchers an almost alchemical ability to transform one type of cell into virtually any other type of cell. Instead of hunting for rare cells, researchers could create iPSCs and then differentiate the iPSCs into neurons, skeletal muscle cells, hepatocytes, and so on, enabling studies on impossible-to-obtain human cells, creating an unlimited supply of genetically-identical human cells for tightly-controlled bioproduction and high-throughput screening, and developing novel cell-based therapeutics.
In addition, by providing a better model for human biology and disease than primary cells derived from animal models, human iPSCs can reduce our dependence on the use of animals for research.
The Challenge of iPSCs
However, iPSCs have not had as significant an impact on our understanding of human biology or the advancement of human health as many scientists might have hoped for, as the iPSC differentiation process has proven to be technically challenging, labor-intensive, and time-consuming. Few labs can consistently differentiate a variety of iPSC lines into healthy, functional, and pure populations of target cells.
The Technology that Brings the Full Promise of iPSCs to Every Lab
Elixirgen Scientific has developed an iPSC differentiation technology that makes it possible for any lab to quickly and easily generate iPSC-derived cells in as little as 1-2 weeks. Our transcription factor-based approach leaves the genome untouched, maintaining the physiological relevance of the cells, and is consistently successful for the reliable generation of target tissue. Available as differentiation kits that deliver the transcription factors through equally efficient mRNA or non-integrating Sendai virus formats, Elixirgen Scientific’s iPSC differentiation technology puts all the advantages of iPSC-derived cells into the hands of every lab.
Transcription-factor Based iPSC Differentiation
Our technology is based on over 20 years of research conducted in the laboratory of Elixirgen Scientific’s founder and Chief Scientific Officer, Minoru Ko, M.D., Ph.D., at the National Institute of Aging (1998-2011) and Keio University School of Medicine (2012-present).
The approach is conceptually very simple—introduce a specific set of transcription factors to iPSCs/ESCs at the appropriate times and in the appropriate media to promote differentiation into the desired cell type. The power behind Elixirgen Scientific’s approach is the hard work spent identifying the right transcription factors, the right time, and the right media.
The Scientific History of Our iPSC Differentiation Technology
While at the National Institute of Aging, Dr. Ko used mouse ES cells to establish and demonstrate the principles of the transcription factor-based iPSC differentiation approach that Elixirgen Scientific’s scientists use today. His lab also generated a large number of mouse ES cell clones and gene expression profiles that are still in use today and available to the research community through the Coriell Cell Repository.
The research at Keio University extended this work further into the more biologically relevant human ES and iPS cells. These technologies have been exclusively licensed from Keio University to Elixirgen Scientific.
Current and Future
Elixirgen Scientific consistently strives to offer rapid, robust, reproducible, and scalable differentiation methods for diverse cell types, cementing our role as an industry leader in advancing biomedical research. If you're unable to locate a specific cell type or differentiation method, please get in touch. With our comprehensive approach, leveraging the Human Gene Expression Correlation Matrix and our extensive plasmid library, we are well-prepared to develop any cell type differentiation protocol. Contact us to discuss the specifics of your research project.